-
- Downloads
Update 10 files
- /Data Annotation/GO_enrichment/GO_molecular_function.png - /Data Annotation/.gitkeep - /Data Annotation/GO_enrichment/GO_cellular_component.png - /Data Annotation/GO_enrichment/GO_biological_function.png - /Data Annotation/GO_enrichment/biomart_for_annotation.R - /Data Annotation/GO_enrichment/GO_enrichment.R - /Data Annotation/GO_enrichment/GO molecular function.png - /Data Annotation/GO_enrichment/GO biological function.png - /Data Annotation/GO_enrichment/GO cellular component.png - /Data Annotation/GO_enrichment/mrna.csv
Showing
- Data Annotation/.gitkeep 0 additions, 0 deletionsData Annotation/.gitkeep
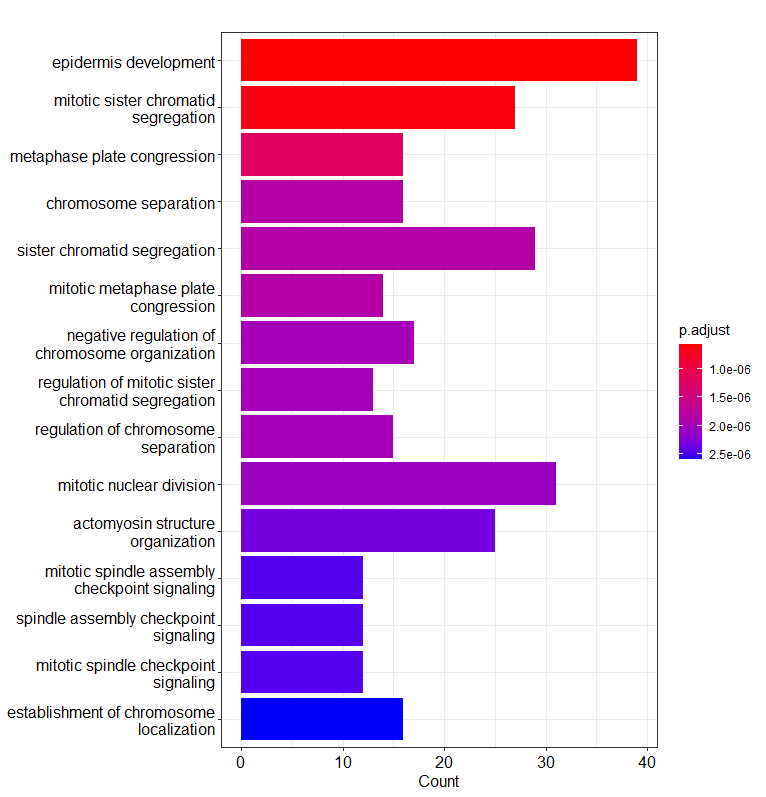
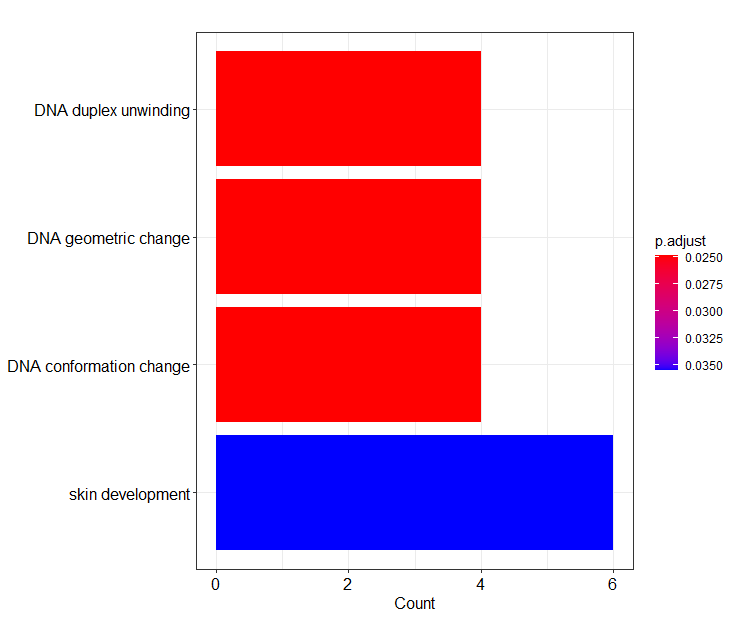
- Data Annotation/GO_enrichment/GO biological function.png 0 additions, 0 deletionsData Annotation/GO_enrichment/GO biological function.png
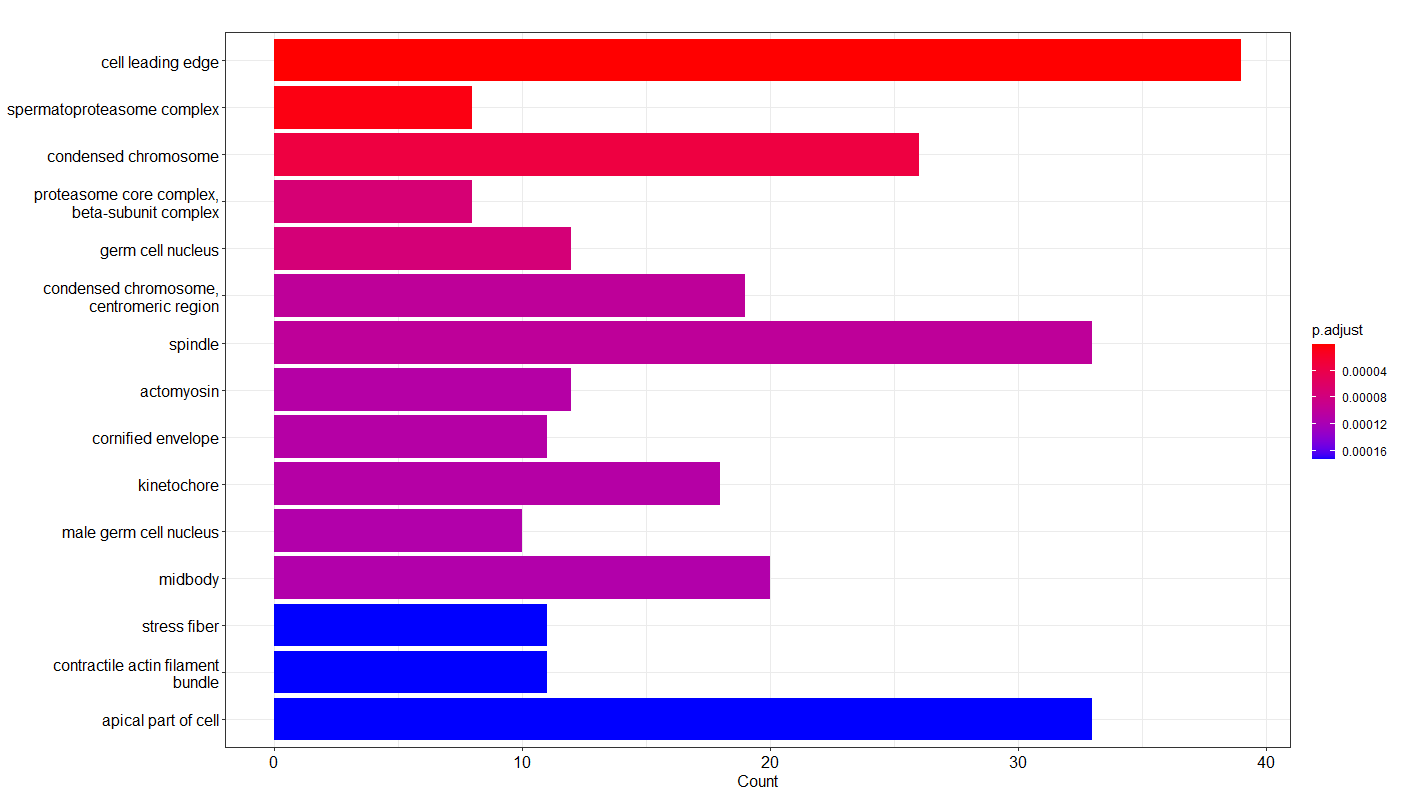
- Data Annotation/GO_enrichment/GO cellular component.png 0 additions, 0 deletionsData Annotation/GO_enrichment/GO cellular component.png
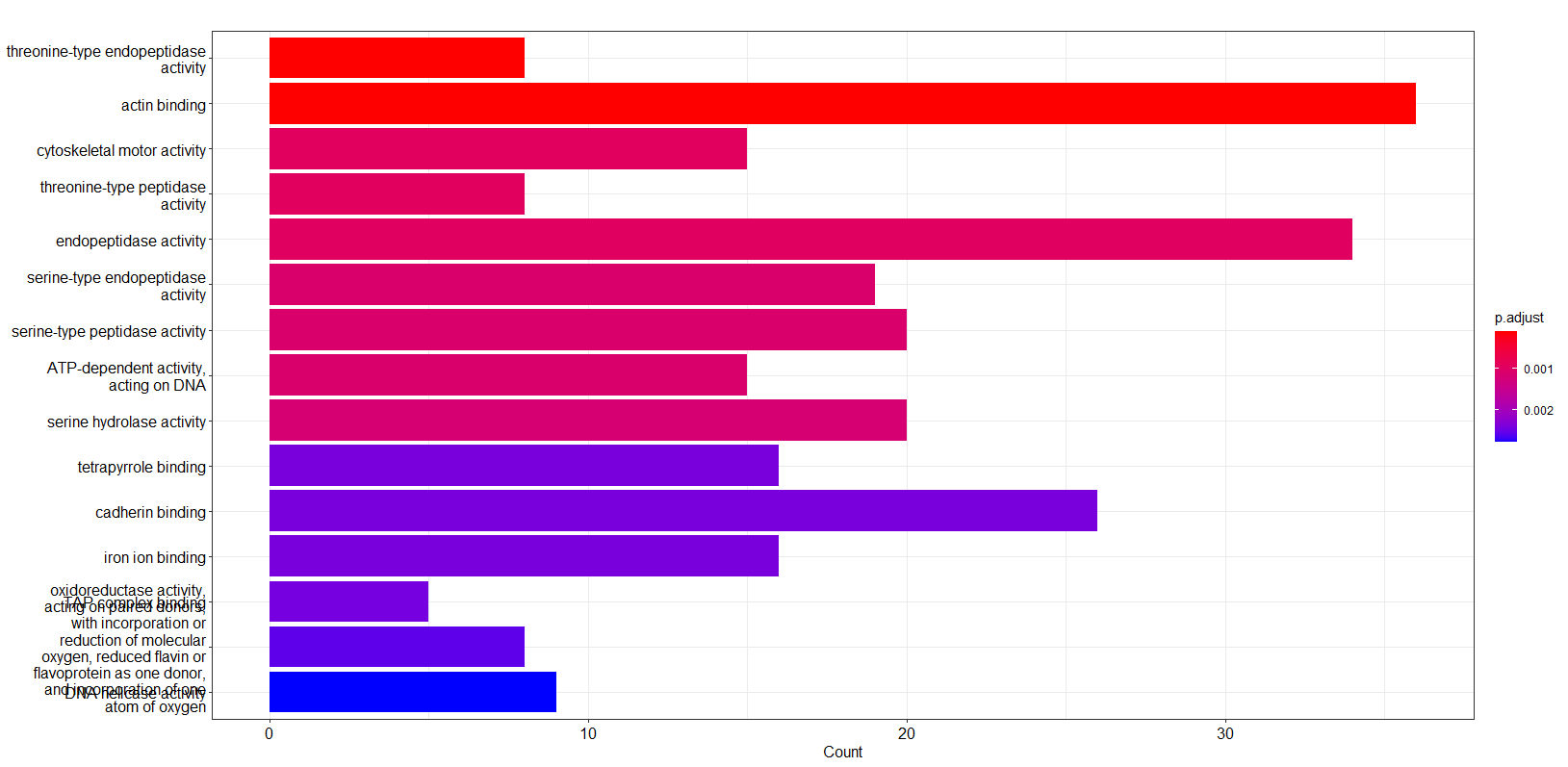
- Data Annotation/GO_enrichment/GO molecular function.png 0 additions, 0 deletionsData Annotation/GO_enrichment/GO molecular function.png
- Data Annotation/GO_enrichment/GO_biological_function.png 0 additions, 0 deletionsData Annotation/GO_enrichment/GO_biological_function.png
- Data Annotation/GO_enrichment/GO_cellular_component.png 0 additions, 0 deletionsData Annotation/GO_enrichment/GO_cellular_component.png
- Data Annotation/GO_enrichment/GO_enrichment.R 2 additions, 5 deletionsData Annotation/GO_enrichment/GO_enrichment.R
- Data Annotation/GO_enrichment/GO_molecular_function.png 0 additions, 0 deletionsData Annotation/GO_enrichment/GO_molecular_function.png
- Data Annotation/GO_enrichment/biomart_for_annotation.R 9 additions, 8 deletionsData Annotation/GO_enrichment/biomart_for_annotation.R
- Data Annotation/GO_enrichment/mrna.csv 579 additions, 0 deletionsData Annotation/GO_enrichment/mrna.csv
Data Annotation/.gitkeep
deleted
100644 → 0
16.9 KiB
19.1 KiB
35.5 KiB
8.26 KiB
11.4 KiB
13.4 KiB
Data Annotation/GO_enrichment/mrna.csv
0 → 100644
This diff is collapsed.